Welcome back to another post from [VS]Codes! Today I’m excited to share another scientific publication that came out of my PhD in biomedical informatics from the University of Pennsylvania, this time in collaboration with the Health Futures team at Microsoft Research (MSR): “Addressing biomedical data challenges and opportunities to inform a large-scale data lifecycle for enhanced data sharing, interoperability, analysis, and collaboration across stakeholders”.
This paper spun out of the project I completed as a “Health AI UX Research Intern” with MSR Health Futures in the summer of 2022 under the mentorship of Mandi Hall - our study explored critical challenges in the biomedical data lifecycle and offered actionable recommendations to improve data sharing, interoperability, and reproducibility across research institutions, healthcare settings, and industry partners.
I have always been passionate about the concept of simplifying medical discovery from bench to bedside, so this project felt like a perfect application of my interests at a scale outside of the work that I had otherwise been pursuing in my PhD. It was a true joy to be able to bring together an interdisciplinary team of experts in AI, biomedical informatics, computational biology, and data science to better understand the pain points that researchers, clinicians, and data professionals encounter when working with complex biomedical datasets.
You can read the official blog post from Microsoft Research summarizing our paper here, as well as the full text of our publication here. With that, let’s dive into a brief overview of the takeaways of our work.
Challenges in the biomedical data lifecycle
Biomedical research is increasingly driven by large-scale, heterogeneous (a.k.a. ‘multiomic’) datasets, with the goal of accelerating precision medicine and improving patient outcomes. However, in spite of numerous advances in the broader fields of machine learning and data science, researchers still face major challenges when handling biomedical data.
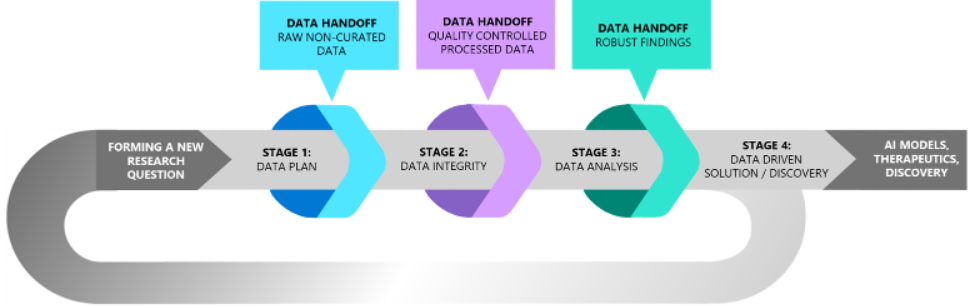
To address these barriers, our study involved in-depth, semi-structured interviews with professionals placed across the biomedical research spectrum, including bench scientists, computational biologists, clinicians, and data curators, with the goal of identifying common pain points and potential solutions. The challenges we discovered included:
Identifying and procuring the appropriate data for a given research question
Curating and validating procured data for downstream analysis
Learning how to apply new analysis methods to validated data and navigating inconsistent computational environments
Distributing data-driven findings effectively and reproducibly
Managing the flow of data across phases of the data lifecycle
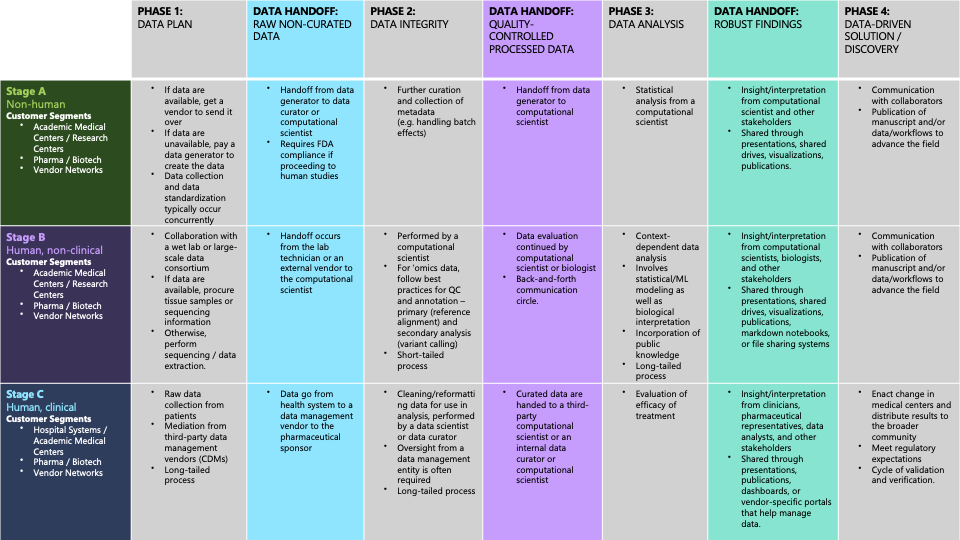
Ultimately, across the challenges identified for biomedical discovery, participant interviews echoed a single message: the significance of collaboration and trust surrounding the flow of data. Each exchange of data involved multiple professional stakeholders, including data generators, research scientists, data curators, third-party vendors, bioinformaticians, computational biologists, biologists, and clinicians. Insight and interpretation are continually needed from all stakeholders involved to ensure the accuracy and integrity of the data.
Key recommendations
Our research highlights the need for a unified approach to the biomedical data lifecycle, from data acquisition to analysis and dissemination. Some of our core recommendations based upon identified challenges included:
- Creating user-friendly platforms that transition from manual data collection to electronic, structured systems.
- Standardizing analysis workflows to improve reproducibility, incorporating version control and seamless integration of computational notebooks.
- Leveraging AI and automation to streamline data ingestion, validation, and processing for unstructured biomedical datasets.
- Building secure, cloud-based infrastructures to facilitate real-time collaboration among researchers across institutions.
By implementing these strategies, we can work toward a more integrated, scalable, and efficient biomedical data ecosystem - one that enables faster discoveries, more reproducible research, and better patient outcomes.
Takeaways
At its core, our study underscores a fundamental truth in biomedical data science: research is only as strong as the data infrastructure supporting it. If we want to harness the full potential of multiomic data, AI-driven analysis, and precision medicine, it is essential that we break down silos, standardize workflows, and rethink how we share and collaborate on data.
I’m incredibly grateful to my co-authors from Microsoft Research and the University of Pennsylvania for making this work possible, and I hope these findings spark further discussions on how we can improve biomedical data practices for the entire research community.